Absence of Clinical Value of TZAP Mutation and Expression in Non-small Cell Lung Cancers
Article information
Abstract
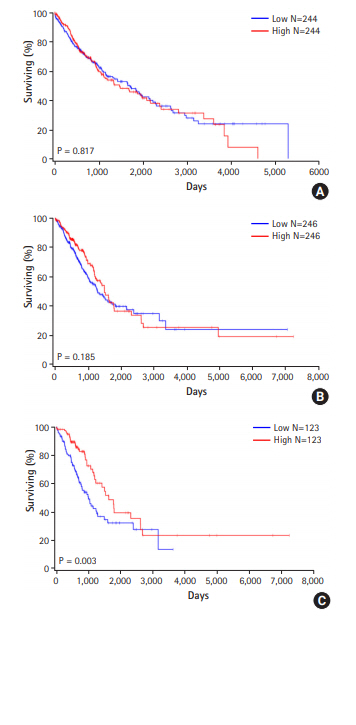
The zinc finger protein ZBTB48 is a telomere-associated factor and renamed it as telomeric zinc finger-associated protein (TZAP). It binds preferentially to long telomeres competing with TRF1 and TRF2. However, its mutation in cancers has not been studied. In the present study, we analyzed TZAP mutation in 134 non-small cell lung cancers (NSCLCs). And its big data analysis was performed using COSMIC and TCGA data analysis. TZAP mutation was not found in 134 NSCLCs. And big data also showed that TZAP mutation was extremely low (0.59%, 15/2548). TCGA survival analysis showed no prognostic value of TZAP expression in lung adenocarcinoma (p = 0.185) and squamous cell carcinoma (p = 0.817). When stratified patients sorting as 25:25 (quarter), it has a significance (p = 0.003). This result suggested that genetic change of TZAP did not appear to be a possible molecular marker in lung cancer.
Introduction
Telomeres, composed of 6-bp TTAGGG repeat sequences, are nucleoprotein complexes capping each end of eukaryotic chromosomes [1,2]. In normal human somatic cells, telomeres have an average length of 5 to 15 kilobases and are shortened by approximately 30 to 200 base pairs at every cell division. Telomere shortening is counteracted by the reverse transcriptase telomerase in stem cells and most types of cancer, while the remaining cancers maintain telomeres with alternative lengthening of telomeres (ALT) mechanism [3-5]. A telomere trimming mechanism is induced in the presence of overly long telomeres, which are cut back to normal length by rapid telomere shortening [6-8]. Though the specific regulation of this process has not been identified, recent studies have discovered a special protein that is necessary for regulating telomere [9]. They identified the zinc finger protein ZBTB48, as a telomere-associated factor, and renamed it telomeric zinc finger-associated protein (TZAP). It binds preferentially to long telomeres competing with telomeric repeat factor 1 and 2 (TRF1 and TRF2). In addition, the study showed that overexpression of TZAP caused progressive telomere shortening [9]. TZAP localizes to chromosome 1p36, a region that is frequently rearranged or deleted in various cancers [10-12]. This genetic change of TZAP may be associated with cancer pathogenesis, however, a genetic analysis of TZAP has not been performed in any specific type of cancer.
Lung cancer is the leading cause of cancer-related deaths worldwide, with only 16.8% of lung cancer patients alive 5 years after diagnosis [13]. Many causative factors for lung cancer have been identified, including active smoking, secondhand smoke, occupational agents, radiation, and environmental pollutants [14-15]. These factors may influence the telomere regulation in human inducing various disease such as especially in nonsmall cell lung cancer (NSCLC). These previous studies suggested that the regulation of telomere length had the potential to serve as prognostic marker for patients with NSCLC, and may therefore aid clinicians in making informed therapeutic decision [16,17]. Therefore, we analyzed TZAP mutation and expression in patients with NSCLC for the first time. To increase the statistical significance, we confirmed big cohorts, The Cancer Genome Atlas (TCGA) and Catalogue of Somatic Mutation In Cancer (COSMIC) data.
Material and Methods
Patients and tissue samples
A total of 134 cases were identified from pathology reports of patients who underwent conventional surgery for NSCLC at the Kyungpook National University Hospital (KNUH). All materials derived from the National Biobank of Korea, KNUH, were obtained under institutional review board‑approved protocols (Approval No. KNUMCBIO_14‑1010). None of the patients received chemotherapy or radiotherapy prior to surgery. We collected basic clinical data including age, gender, disease stage, and smoking status. The pathologic staging of lung cancer was based on the 7th AJCC staging system.
DNA samples of the tumor tissue and adjacent normal mucosa were obtained using an extraction kit according to the manufacturer’s instructions (Absolute™ DNA extraction Kit, BioSewoom, Korea). The quantity and quality of DNA were measured using NanoDrop 1000 (Thermo Fisher Scientific, Inc., Pittsburgh, PA, USA).
TZAP mutation analysis
TZAP exons were amplified from isolated DNA using the polymerase chain reaction (PCR). PCR was performed using AmpliTaq Gold® DNA Polymerase (Applied Biosystems, Thermo Fisher Scientific, Inc., Waltham, MA, USA). The sequences of primers for all exons are presented in Table 1. Thermocycling was conducted under the following conditions: 40 cycles of 94°C for 30 sec, 55-57°C for 30 sec, and 72°C for 60 sec. The PCR products were separated on a 1.5% agarose gel using electrophoresis and stained with ethidium bromide for 20 min to confirm the size of the bands. Direct DNA sequencing of the TZAP was subsequently performed using an ABI 3730 DNA sequencer (Bionics Inc., Seoul, South Korea).
The Cancer Genome Atlas (TCGA) data analysis
To investigate the clinical significance of TZAP, we used the TCGA and COSMIC database. TZAP mRNA expression data were downloaded from TCGA’s data portal (https://tcga-data.nci.nih.gov/tcga/) on October, 2019 [18]. And TZAP mutation data were checked form COSMIC (https://cancer.sanger.ac.uk/cosmic) on October, 2019 [19]. Its clinicopathological and prognostic values were analyzed.
Statistical analysis
Statistical analysis was performed by SPSS version 23.0 (IBM SPSS, Armonk, NY, USA). Survival curves, constructed using the univariate Kaplan-Meier estimators, were compared using the log-rank test. Overall survival (OS) was defined as the time between diagnosis and mortality. A p value of <0.05 denoted significance for all statistical analyses performed in this study.
Results
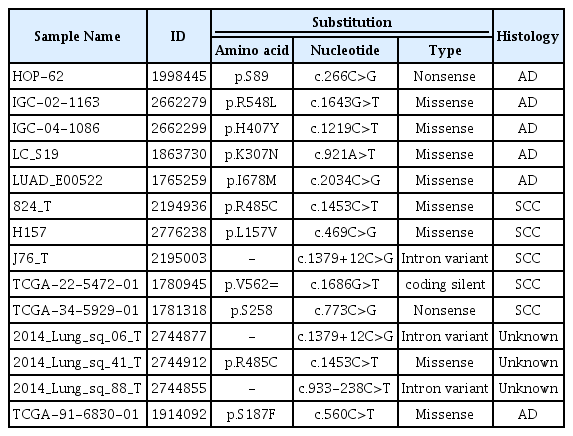
The sequences of TZAP regions were successfully analyzed in the 134 NSCLCs tissue samples. We found no TZAP mutations in all NSCLCs. In COSMIC data, this mutation was found in 0.58% (15/2548) of NSCLC. It was summarized in Table 2. Most mutations were missense mutation and it did not have any clinical significance.
We then assessed the survival analysis to clarify the prognostic significance of the TZAP expression in patients with NSCLC using TCGA data. In 488 squamous cell carcinomas, TZAP expression did not show any prognostic value for the patients with NSCLCs (p = 0.817, Fig. 1A). In 246 adenocarcinomas, TZAP also did not have any prognostic significance (p = 0.185, Fig. 1B). However, when the patients were divided by quarter, poorer survival results were found in lower expression group (p = 0.003, Fig. 1C).
Discussion
In this study, we demonstrated TZAP mutation and expression for the first time in cancer, specifically NSCLC. TZAP mutations have not yet been studied and rare mutations were only reported in the International Cancer Genome Consortium (ICGC) data [19]. In our hospital samples, we found no TZAP mutation in lung cancer and non-cancerous tissue, respectively. COSMIC data also reported an extremely lower frequency of TZAP mutation in lung cancers. Thus, this mutation may be not specific for lung cancer, although this needs to be further confirmed.
ZBTB48 is a Kruppel-like C2H2 zinc finger protein consisting of 11 tandem zinc finger domains located C-terminal to the BTB/POZ domain [9]. Recent study established that ZBTB48, renamed by Li et al. [9] as TZAP for telomeric zinc finger-associated protein, tend to bind overly long telomeres in mouse stem cells and cancer cells, irrespective of whether telomerase or ALT is operational. TZAP appears to compete with TRF1 and TRF2 for binding to telomeres, and its association could be contingent on the exhaustion of free shelterin, possibly as telomeres replicate and elongate [8,9]. Therefore, its mutation or expression may be associated with various diseases, especially cancers. Based on this hypothesis, we searched ZBTB48 (TZAP) expression level in TCGA data and analyzed its prognostic value in lung cancers.
According to the TCGA data, TZAP expression, unfortunately, was not associated with lung cancer survival. However, it showed a possibility for prognostic value in some classification. Our study has demonstrated, for the first time, a better prognosis in patients with NSCLC with higher TZAP expression. However, TZAP did not have any clinical and prognostic values totally. Considering the important role of telomere regulation in lung cancer [12,16,17], precise mechanism of genetic change of TZAP should be studied further.
In conclusion, we studied the TZAP mutation and TZAP expression in patients with NSCLC. TZAP did not have great significance for clinical and prognostic markers in NSCLC. The results of the present study warrant future large-scale studies to elucidate the underlying molecular mechanisms of TZAP and to determine the potential clinical utility.
Notes
Conflict of interest
All authors declare no conflicts-of-interest related to this article.