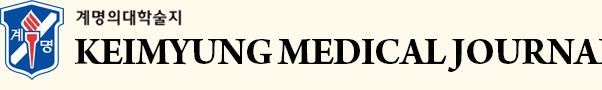|
 |
- Search
| Keimyung Med J > Volume 41(1); 2022 > Article |
|
Abstract
DNA methyltransferase 3B (DNMT3B), one of DNA methyltransferases has many roles in DNA methylation and cancer pathogenesis. However, its clinical and prognostic value was not studied in hepatocellular carcinoma (HCC). In this study, we analyzed DNMT3B expression in HCC by public big data, The Cancer Genome Atlas. Primary data about total 360 HCC were downloaded and its clinicopathological implication was analyzed. M stage (p = 0.07) and pathologic stage (p = 0.04) were associated with DNMT3B expression, though M stage did not get a statistical significance. And alpha fetoprotein level was positively correlated with DNMT3B expression (p < 0.001). Higher DNMT3B expression predicted shorter overall survival in HCC patients (p < 0.05). Disease-free survival was shorter in HCC with lower DNMT3B expression, within borderline statistical significance (p = 0.081). It suggested that DNMT3B expression may have an important role in HCC prognosis and its detail mechanism should be confirmed further.
De novo methylation is occurred frequently during the early embryogenesis and is faithfully copied following DNA replication at each cell cycle [1]. DNA methylation is involved in many embryonic developments, biological processes, and cell differentiation [2]. The dysregulation of DNA methyltransferases (DNMTs) and subsequent aberrant DNA methylation is a key feature of human malignancies [3,4]. DNMT1, DNMT3A, and DNMT3B are the enzymatic players of DNA methylation [1,2]. DNMT3B overexpression is frequently found in tumors, especially in 30% of breast cancers [5,6]. And it result into variation in the targeting efficiency and abnormal catalytic activity contributing to cancer development and progression. Therefore, DNMT3B may act as an oncogene, and its overexpression induces an unfavorable prognosis [3-6].
Hepatocellular carcinoma (HCC) is a majority of human cancers in worldwide, and is a leading cause of death in Korea [7,8]. Recent studies about HCC showed an alteration of DNA methylation by dysregulation of DNMT3B [9,10]. Yu et al. described that telomerase reverse transcriptase (TERT) regulates DNMT3B expression in HCC and their co-operation may predict a poorer prognosis [11]. Recent advances in genomic profiling using next-generation sequencing have made it possible to identify the genetic characteristics of cancer. Large-scale cancer genome studies such as The Cancer Genome Atlas (TCGA) used to investigate genes in different cancer types [12]. However, clinicopathological characteristics have not been studied in HCC. Therefore, we aimed to examine the clinicopathologic and prognostic value of DNMT3B expression in HCC using gene expression RNA sequencing (RNAseq) data obtained from TCGA datasets.
Primary data were downloaded from TCGA data portal in March 2022. TCGA dataset consisted of 360 samples, including primary HCCs and adjacent tissues. The RNAseq data of HCC were sorted from TCGA with DNMT3B mRNA expression and clinical parameters. This study met the publication guidelines for using TCGA datasets (http://www.cancer.gov/about-nci/organization/ccg/research/structrual-genomics/tcga/using-tcga/citing-tcga). Overall survival (OS) was defined as the days from the date of surgery to the date of the last follow-up visit or the date of death due to any cause. And disease-free survival (DFS) was defined as the days from surgery to any type of recurrence.
The Statistical Package for the Social Sciences (SPSS), version 24.0 for Windows (IBM, Armonk, NY, USA), was used for all statistical analysis. Chi-square and Mann-Whitney U-tests were used to analyze the relationship between variables. For survival analysis, the mean gene expression was used as a cutoff to divide the patients into high- and low-expression groups. Survival analysis was performed using the Kaplan–Meier method, and the log rank test was used to identify statistically significant differences between the two groups. A two-tailed P value < 0.05 was considered to signify statistical significance.
Clinicopathological characteristics of DNMT3B mRNA expression were analyzed from TCGA data and presented in Table 1. Patients were divided into two groups according to the expression level of DNMT3B, and their clinical features were statistically analyzed. Age and sex were not different according to the DNMT3B expression level. Metastasis and Stage IV cancers were found in only lower expression of DNMT3B. Therefore, M stage (p = 0.07) and pathologic stage (p = 0.04) were associated with DNMT3B expression, though M stage did not get a statistical significance. And serum alpha fetoprotein (AFP) level was positively correlated with DNMT3B expression (p < 0.001).
The median follow-up period in the cohort examined in the survival analysis was 2032 ± 117 days (range: 9–3478 days). Univariate survival analysis revealed that shorter OS in HCC patients was associated with higher DNMT3B expression (1840.26 ± 164.36 vs. 2180.06 ± 150.16 days, p = 0.003, Fig. 1A). DFS was shorter in HCC patients with lower DNMT3B expression, it did not get a statistical significance (1474.18 ± 153.43 vs. 1625.30 ± 165.52 days, p = 0.081; Fig. 1B).
Cancer is a kind of genetic disease, however, recent study introduced that epigenetic change are closely involved in this process. Therefore, to understand DNA methylation, one of the main events of epigenomic patterns, is extremely important in cancer. The DNA methylation is catalyzed by methyltransferases DNMT3A and DNMT3B, however, exact role of DNMT3B in cancer is not fully understood.
For a first time, we demonstrated clinical value of DNMT3B expression in HCC. Previous study demonstrated that DNMT3B expression level was remarkably higher in HCC than in non-tumorous tissue [13]. And this study showed DNMT3B deletion induced liver carcinogenesis suggesting its protective role against liver inflammation and HCC development. Many studies focused oncogenic role of DNMT3B in cancer [6,9,10] and our survival analysis also found that higher expression of DNMT3B predicts poorer prognosis. And its expression was positively correlated to AFP level.
However, its clinical implication was related to favoring prognosis. Our data showed that lower expression of DNMT3B was found only in metastasis and stage IV cancers. This clinical character of DNMT3B expression may be associated with its protective role in HCC, and it agreed with these result [14-16]. These data suggested that lack of DNMT3B facilitated cancer progression suggesting that its deficiency induced genetic instability. These completely opposite findings suggest that DNMT3B may have a paradoxical effect in cancer, as an oncogene and as a tumor suppressor gene. A recent study showed that higher TERT and DNMT3B expressions had statistically poorer survival in HCC patients by TCGA data [11]. And they suggested that TERT-DNMT3B-PTEN-AKT axis has multi-oncogenic activities in cancers. Therefore, the authors suggested that the role of DNMT3B in cancer should be confirmed in a larger scale study further. Then, its molecular mechanism should be identified, and prognostic value of DNMT3B in various cancers should be confirmed.
Here, we revealed clinical significance of DNMT3B mRNA expression in HCC by public data. DNMT3B expression may have various roles in hepatocellular carcinogenesis and future study about molecular mechanisms of DNMT3B will be required.
Fig. 1.
Survival analysis of DNA methyltransferase 3B (DNMT3B) expression in hepatocellular carcinomas. (A) Overall survival. (B) Disease free survival.
Table 1.
The Cancer Genome Atlas data of DNMT3B expressions in HCC
References
1. Iurlaro M, von Meyenn F, Reik W. DNA methylation homeostasis in human and mouse development. Curr Opin Genet Dev. 2017;43:101–9.


2. Leppert S, Matarazzo MR. De novo DNMTs and DNA methylation: novel insights into disease pathogenesis and therapy from epigenomics. Curr Pharm Des. 2014;20:1812–8.


3. Bishop KS, Ferguson LR. The interaction between epigenetics, nutrition and the development of cancer. Nutrients. 2015;7:922–47.



4. Duan F, Cui S, Song C, Dai L, Zhao X, Zhang X. Systematic evaluation of cancer risk associated with DNMT3B polymorphisms. J Cancer Res Clin Oncol. 2015;141:1205–20.


5. Palakurthy RK, Wajapeyee N, Santra MK, Gazin C, Lin L, Gobeil S, et al. Epigenetic silencing of the RASSF1A tumor suppressor gene through HOXB3-mediated induction of DNMT3B expression. Mol Cell. 2009;36:219–30.



6. Hayette S, Thomas X, Jallades L, Chabane K, Charlot C, Tigaud I, et al. High DNA methyltransferase DNMT3B levels: a poor prognostic marker in acute myeloid leukemia. PLoS One. 2017;7:DOI: 10.1371/journal.pone.0051527.

7. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33.


8. Vauthey JN, Lauwers GY, Esnaola NF, Do KA, Belghiti J, Mirza N, et al. Simplified staging for hepatocellular carcinoma. J Clin Oncol. 2002;20:1527–36.


9. Fan H, Zhao Z, Cheng Y, Cui H, Qiao F, Wang L, et al. Genome-wide profiling of DNA methylation reveals preferred sequences of DNMTs in hepatocellular carcinoma cells. Tumour Biol. 2016;37:877–85.


10. Oh BK, Kim H, Park HJ, Shim YH, Choi J, Park C, et al. DNA methyltransferase expression and DNA methylation in human hepatocellular carcinoma and their clinicopathological correlation. Int J Mol Med. 2007;20:65–73.


11. Yu J, Yuan X, Sjöholm L, Liu T, Kong F, Ekström TJ, et al. Telomerase reverse transcriptase regulates DNMT3B expression/aberrant DNA methylation phenotype and AKT activation in hepatocellular carcinoma. Cancer Lett. 2018;434:33–41.


12. Cancer Genome Atlas Research Network. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169:1327–41.e23.


13. Iguchi E, Takai A, Takeda H, Kumagai K, Arasawa S, Eso Y, et al. DNA methyltransferase 3B plays a protective role against hepatocarcinogenesis caused by chronic inflammation via maintaining mitochondrial homeostasis. Sci Rep. 2020;10:DOI: 10.1038/s41598-020-78151-2.

14. Vasanthakumar A, Lepore JB, Zegarek MH, Kocherginsky M, Singh M, Davis EM, et al. Dnmt3b is a haploinsufficient tumor suppressor gene in Myc-induced lymphomagenesis. Blood. 2013;121:2059–63.



-
METRICS

-
- 2 Crossref
- 2,188 View
- 24 Download
- Related articles in Keimyung Med J
-
A Case of Cutaneous Metastasis to the Scalp from Hepatocellular Carcinoma2004 December;23(2)